Streamlining Healthcare: Fine-Tuning LLMs to Save Time for Medical Personnel
Discover how we fine tuned an open source LLM for a domain specific medical use case.
written by Jacob Owens - 11/20/2023
Read time: 11 minutes
The fascination with fine-tuning large language models (LLMs) has surged recently, sparking widespread interest in their potential applications. Like many others, we were captivated by this potential and eager to explore the limits and capabilities of what LLMs can achieve. Our problem was relatively simple, we spend dozens of hours per week writing and interpretating Electromyography/Nerve conduction study (EMG/NCS) tests. The goal for our project: to mostly automate the tedious process of generating reports for tests; drawing on patient history, physical examination, height, and NCS/EMG data. After a week or so of trial and error, the results were pretty impressive. Not only did our custom LLM streamline the report generation process, but it also opened up a new realm of possibilities in medical diagnostics and patient care. This blog post delves into the exciting journey where technology intersects with healthcare, exploring how AI is not just assisting but revolutionizing our approach to medicine.
Curating the dataset
A few facts about the dataset:
- The dataset we used includes more than 21,000 tests conducted over a span of 20 years across multiple locations and 4 machines.
- Exams were performed on a diverse set of individuals ranging from age 12 - 95.
- Motor, Sensory, F-wave, H-reflex, and EMG results were used in training.
- No Facial or repetitive stim results were used in training.
- No PHI was included in the dataset. Only the raw test results as well as de-identified physical exam and history were used in training.
This is great and all, but there was a few problems that needed to be addressed before I could do anything with any of the data.
All test results were stored in Microsoft Word documents. At first I thought this would not be a big deal. I could use the docx python library to programatically loop through each document and extract the relevant tables. Except, this was Microsoft Word 1997, so these were not .docx files but .doc files (2 different things entirely). So I needed to programatically save each one as a .docx file first. I did some research and discovered I could use the pywin32 library to control Word programatically on my PC. This process took about 3.5 seconds for each word document, so 3.5 sec x ~20,000... you do the math.
import os
import win32com.client as win32
import time
def convert_doc_to_docx(doc_path, save_dir):
word = win32.Dispatch('Word.Application')
word.Visible = False # Do not open the application window
docx_path = os.path.join(save_dir, os.path.basename(doc_path) + "x") # Add 'x' to make it .docx
try:
doc = word.Documents.Open(doc_path)
doc.SaveAs(docx_path, FileFormat=16) # FileFormat=16 for docx
except Exception as e:
print(f"Error converting file {doc_path}: {e}")
finally:
doc.Close()
word.Quit()
time.sleep(1) # Sleep for 1 second to give Word time to close before deleting the object
folder_path = "C:\\path" # Replace with your folder path
save_dir = 'C:\\other\path' # New directory to save converted files
if not os.path.exists(save_dir):
os.makedirs(save_dir)
start_time = time.time()
for file in os.listdir(folder_path):
if file.endswith(".doc"):
doc_path = os.path.join(folder_path, file)
convert_doc_to_docx(doc_path, save_dir)
elapsed_time = time.time() - start_time # Time taken for this file
print(f"Converted {file}. {elapsed_time:.2f} seconds elapsed.")
end_time = time.time() # End the timer
elapsed_time = end_time - start_time # Calculate the elapsed time
print(f"Conversion completed in {elapsed_time} seconds.")
After this process completed (19.5 hours later...) the files were ready to process.
Processing the dataset
I wrote the script below to loop through each file, extract the bits I needed, and save to a csv.
import os
import re
from docx import Document
import pandas as pd
import time
def extract_section(text, start_marker, end_marker=None):
"""Extracts a section of text between two markers."""
if end_marker:
pattern = f"{start_marker}(.*?){end_marker}"
else:
pattern = f"{start_marker}(.*)"
match = re.search(pattern, text, re.DOTALL)
return match.group(1).strip() if match else None
def extract_tables(doc):
"""Extracts all tables from a Word document."""
tables = []
for table in doc.tables:
table_data = []
for row in table.rows:
row_data = [cell.text for cell in row.cells]
table_data.append(row_data)
tables.append(table_data)
return tables
def read_docx(file_path):
"""Reads a Word document and extracts text and tables."""
doc = Document(file_path)
full_text = "\n".join([para.text for para in doc.paragraphs])
# Extracting tables
tables = extract_tables(doc)
# Check if the expected number of tables is present
num_tables = len(tables)
height_str = tables[0][3][1]
height_inches = None
# Parsing and converting height
if height_str:
match = re.search(r"(\d+)\s*feet\s*(\d+)\s*inch", height_str)
if match:
feet = int(match.group(1))
inches = int(match.group(2))
height_inches = feet * 12 + inches
# Extracting specific sections
# history = testHistory if testHistory not in [None, ""] else None
history = tables[0][5][1]
pe = extract_section(full_text, "PE:", "Interpretation:")
interpretation = extract_section(full_text, "Interpretation:", "Conclusion:")
conclusion = extract_section(full_text, "Conclusion:", "NCS performed by:")
motor_ncs = tables[1] if num_tables > 0 else None
sensory_ncs = (
tables[2]
if num_tables > 2 and tables[2][0][1] != "F min"
else (tables[1] if num_tables > 1 else None)
)
f_wave = tables[3] if num_tables > 3 else None
h_reflex = None
emg = None
if num_tables > 4:
if tables[4][0][1] == "H Latency":
h_reflex = tables[4]
emg = tables[5] if num_tables > 5 else None
else:
emg = tables[4]
return {
"filename": os.path.basename(file_path),
"history": history,
"physical_examination": pe,
"interpretation": "Interpretation: " + interpretation
if interpretation is not None
else None,
"conclusion": "Conclusion: " + conclusion if conclusion is not None else None,
"motor_ncs": motor_ncs,
"sensory_ncs": sensory_ncs,
"f_wave": f_wave,
"h_reflex": h_reflex,
"emg": emg,
"tables": tables,
"Height": height_inches,
}
folder_path = "your/folder/path"
extracted_data = []
all_files = [
file
for file in os.listdir(folder_path)
if file.endswith(".docx") and not file.startswith("~$")
]
total_files = len(all_files)
print(f"Detected {total_files} .docx files to process.")
start_time = time.time() # Start time of the entire process
for index, file in enumerate(all_files):
file_path = os.path.join(folder_path, file)
file_start_time = time.time() # Start time for this file
try:
data = read_docx(file_path)
extracted_data.append(data)
except Exception as e:
print(f"Skipping file {file} as it could not be opened.")
continue
finally:
file_end_time = time.time() # End time for this file
file_duration = file_end_time - file_start_time
remaining_files = total_files - (index + 1)
average_time_per_file = (file_end_time - start_time) / (index + 1)
estimated_time_remaining = average_time_per_file * remaining_files
print(f"Processed file {file} in {file_duration:.2f} seconds.")
print(
f"{remaining_files} files remaining. Estimated time remaining: {estimated_time_remaining:.2f} seconds."
)
# Convert to DataFrame for easier manipulation and storage
df = pd.DataFrame(extracted_data)
# Dropping the 'tables' column
df = df.drop("tables", axis=1)
# Saving to CSV without the 'tables' column
df.to_csv("extracted_data.csv", index=False)
print("Done!")Final Cleaning
Finally, I removed all empty, corrupted, or incomplete data from the csv and my dataset was complete.
Choosing an open source LLM
There has been a growing amount of impressive open source LLMs released in the past 6 months. After doing some testing and experimentation with a variety of different models including Llama 2, we decided to fine tune Mistral 7B, a new 7 billion parameter model with remarkable capabilities for its size. Mistral 7B has been shown to be particularly good for fine tuning across many different text generation tasks. The size, speed, and accuracy was more than good enough for our use case.
Fine tuning the model
The model was trained on a single Google Colab A100 GPU for 54 hours using huggingface's autotrain advanced python API. The autotrain advanced API requires a csv file with a single "text" column with a prompt for training. We used the following template for our "text" column.
"###Human:
Analyze the provided patient history, physical exam findings, nerve conduction studies, and EMG results to interpret and conclude the results of the patient's nerve conduction study. Please be aware of the following when interpreting the results: 1. The F-Wave and H-Reflex tests are based on the patient's height in inches. 2. If one nerve's amplitude is less than 50% of the contralateral side, please make a note of that in the report.
- Patient History: {row['history']}
- PE: {row['physical_examination']}
- Height {row['Height']} inches
- Motor Nerve Conduction Findings: {row['motor_ncs']}
- Sensory Nerve Conduction Findings: {row['sensory_ncs']}
- F-Wave Results: {row['f_wave']}
- H-Reflex Results: {row['h_reflex']}
- Electromyography (EMG) Findings: {row['emg']}")
###Assistant:
{row['interpretation']}
{row['conclusion']}
"Then, we used this command to begin the training process and uploading our model to the huggingface hub on completion.
!pip install -q pandas
!pip install -q autotrain-advanced
!autotrain setup --update-torch
from huggingface_hub import notebook_login
notebook_login()
# test csv loaded correctly
import pandas as pd
df = pd.read_csv("train.csv")
df['text'][13]
!autotrain llm --train --project_name project_name --model bn22/Mistral-7B-Instruct-v0.1-sharded --data_path . --use_peft --use_int4 --learning_rate 2e-4 --auto_find_batch_size --num_train_epochs 3 --trainer sft --target_modules q_proj,v_proj --push_to_hub --repo_id repo_id --merge_adapter --block_size 4000 --model_max_length 4000
# we only used such a large block size and model length (4000) because our training inputs were very longTesting the model
Now the fun part, we get to see how well our model performs on our task. For this, I went ahead and set up a testing suite in our application. When an example is selected, the correct Interpretation and conclusion is shown below as a reference to what the model produced. The model was not trained on any of the testing examples.
Let's see how well the model performs on a regular day-to-day polyneuropathy diagnosis.

As you can see, the model performs remarkably close to what a real physician wrote about the interpretation of the test. The only thing it missed was
- "with slight progression since previous nerve studies performed in November 2021" because the model did not have access to know this patient had a previous NCS performed.
Lets try a more difficult example ( a Brachial plexopathy) which is a rare diagnosis and is not seen regularly throughout the dataset.
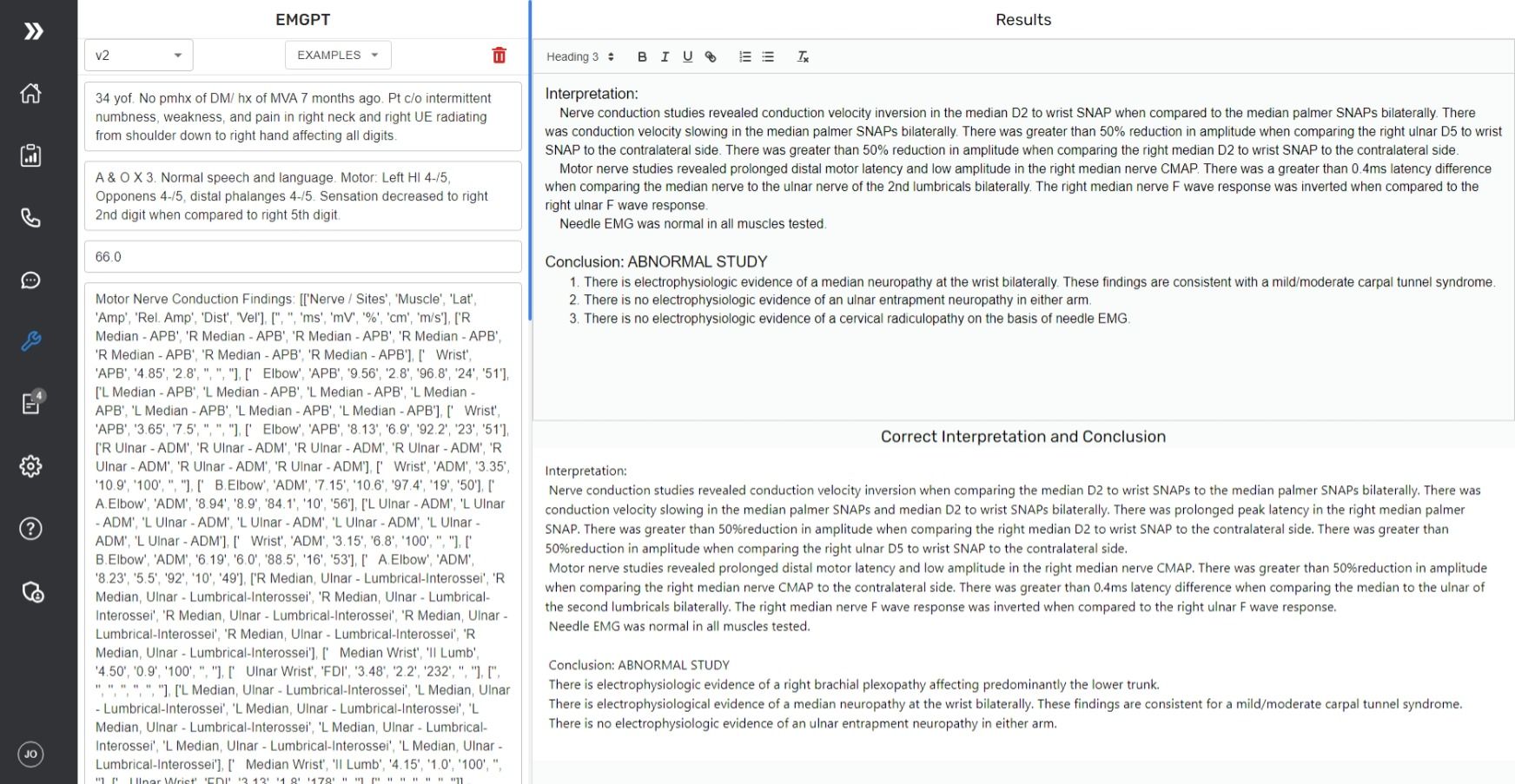
If you compare the correct response and the model response closely, you can tell the model is missing a couple things here.
- It did not conclude the patient had a right brachial plexopathy
- It left out "There was prolonged peak latency in the right median palmer SNAP" in the sensory nerve findings and "There was greater than 50%reduction in amplitude when comparing the right median nerve CMAP to the contralateral side." in the motor nerve findings.
- It did include "There is no electrophysiologic evidence of a cervical radiculopathy on the basis of needle EMG." Which is interesting because it included that by looking back at the patients history which included "pain in right neck and right UE radiating from shoulder down to right hand affecting all digits."
This output is acceptable because the model is not diagnosing the patient, only reducing boilerplate and saving the provider time by outputting 90% correct responses the provider can look over and correct if needed.
We believe we can achieve better results given more training data as well as Reinforcement learning from human feedback (RLHF) in the future.
Conclusion
In conclusion, our experiments with fine-tuning large language models for medical use cases have been an insightful and rewarding journey. From the initial stages of converting Word documents older than most interns into a usable dataset, to the meticulous process of training and fine-tuning the Mistral 7B model, each step has been a learning experience that underscores the immense potential of AI in healthcare. Testing the model highlighted the potential and future prospects of AI in enhancing diagnostic processes.
If you found this article interesting and are looking to leverage AI for a custom medical use case, please contact us to talk about your use case. We would love to collaborate with you to tailor AI solutions that meet your specific needs. We believe the medical field is ripe for technological innovation, and with our expertise, we can forge a path towards more advanced, efficient, and patient-centric healthcare solutions.